
New on bioRxiv: Our collaboration with Jun Ding’s lab at Stanford on thalamic integration during motor learning!
Thalamic integration of basal ganglia and cerebellar circuits during motor learning.
↗ 10.1101/2024.10.31.621388

PKC sensor preprint is now available!
A high-performance genetically encoded sensor for cellular imaging of PKC activity in vivo.
↗ 10.1101/2024.07.19.604387

Dr. Mao has been promoted to Senior Scientist / Professor!
Tianyi Mao has been promoted to Senior Scientist / Professor!
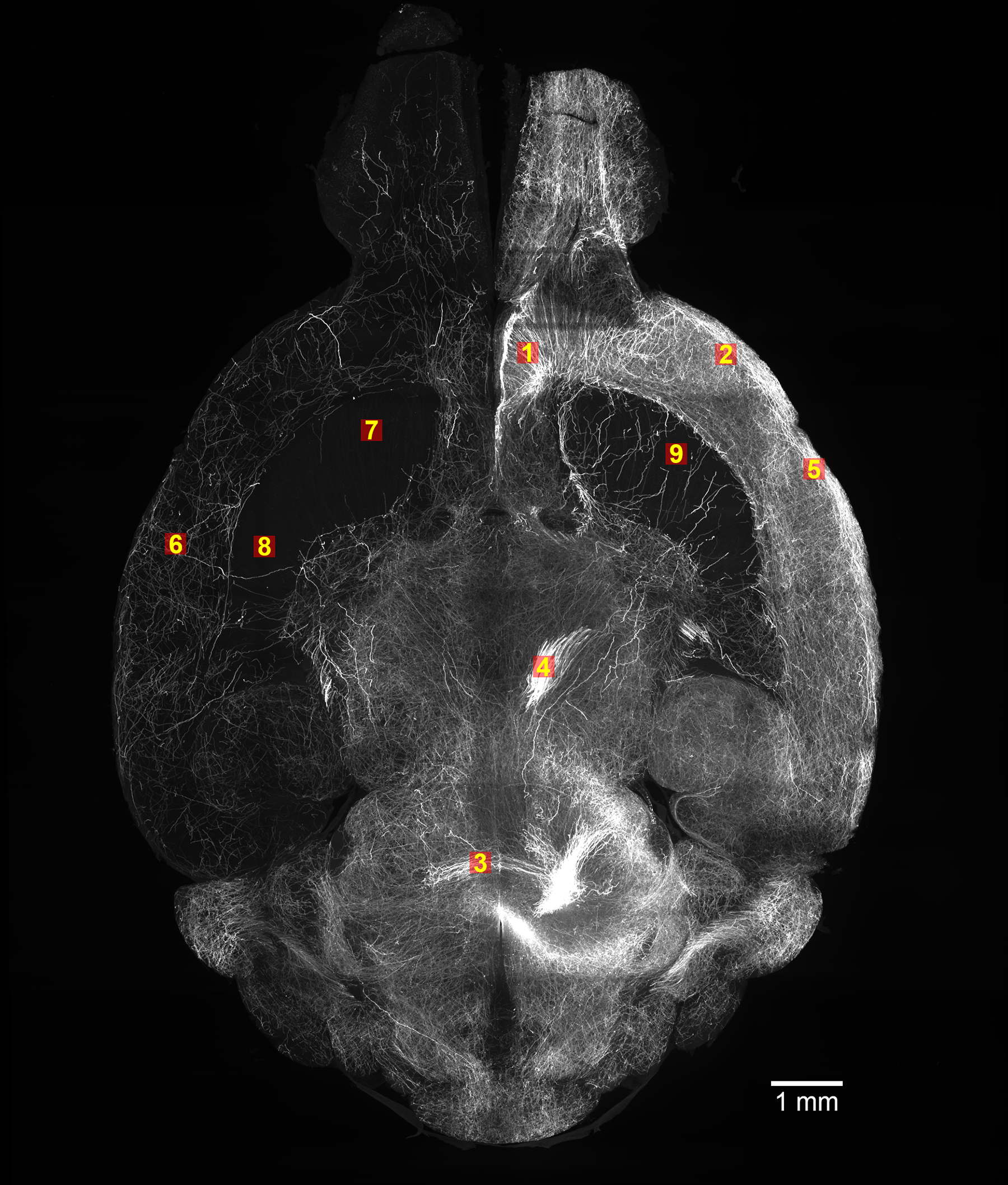
Our collaborative effort with PNNL on deep learning in axon segmentation is out!
Fine-tuning TrailMap: The utility of transfer learning to improve the performance of deep learning in axon segmentation of light-sheet microscopy images.
↗ 10.1371/journal.pone.0293856




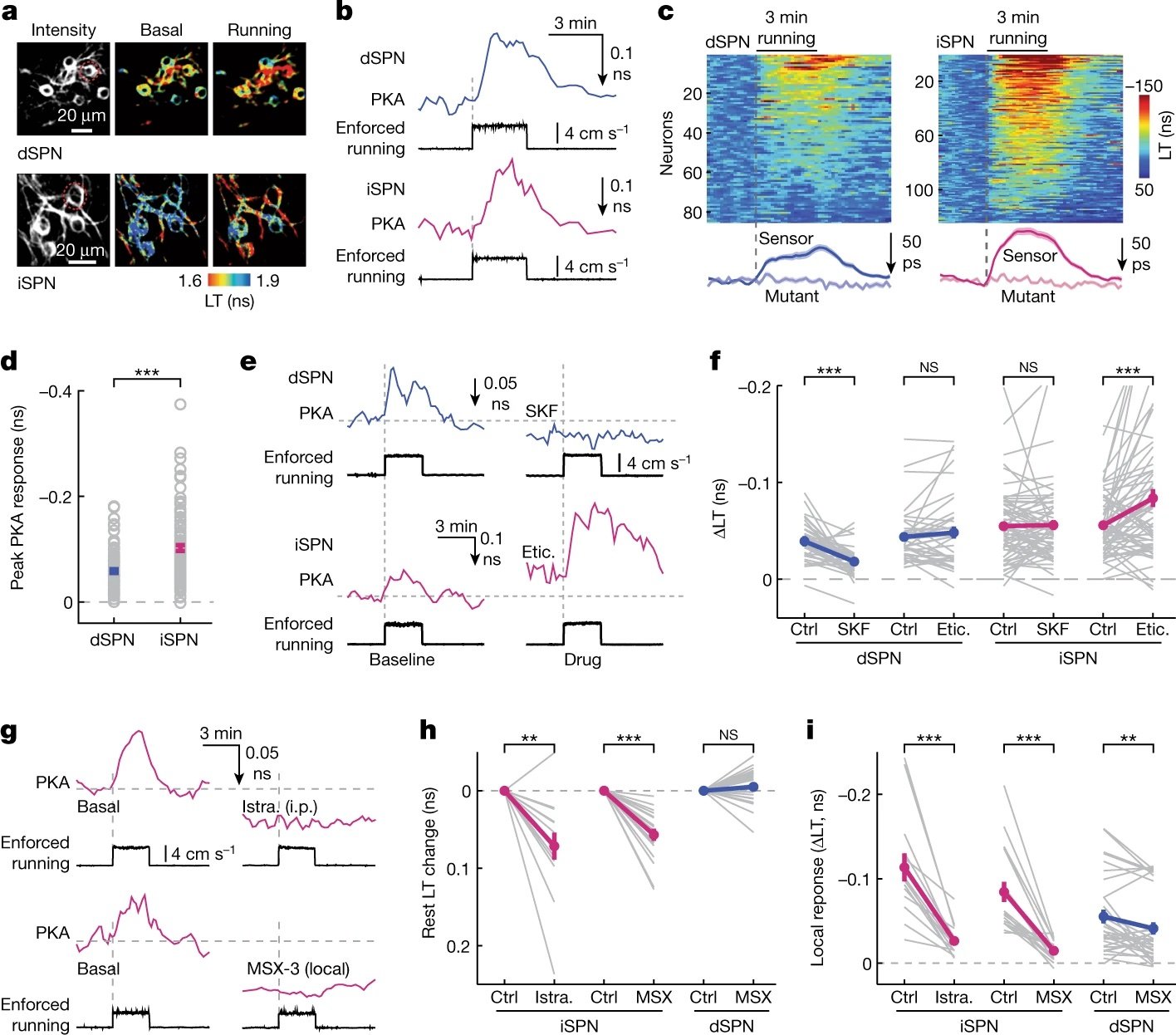
Read our Nature paper revealing an unexpected role for adenosine on PKA signaling in the striatum
Locomotion activates PKA through dopamine and adenosine in striatal neurons.
↗ 10.1038/s41586-022-05407-4
↗ OHSU News
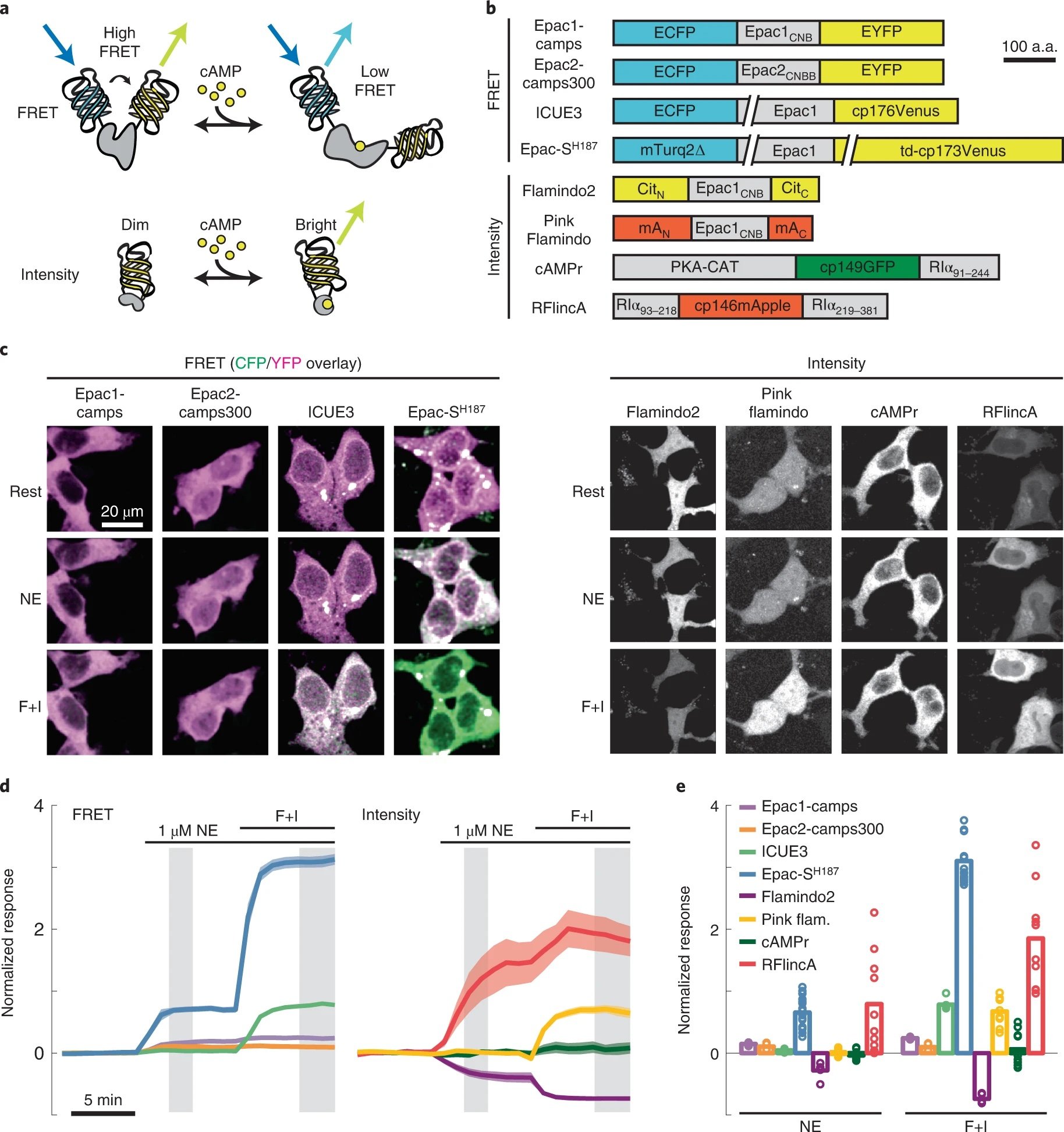
Our cAMPFIRE sensors are now published in Nature Methods!
Sensitive genetically encoded sensors for population and subcellular imaging of cAMP in vivo.
↗ 10.1038/s41592-022-01646-5
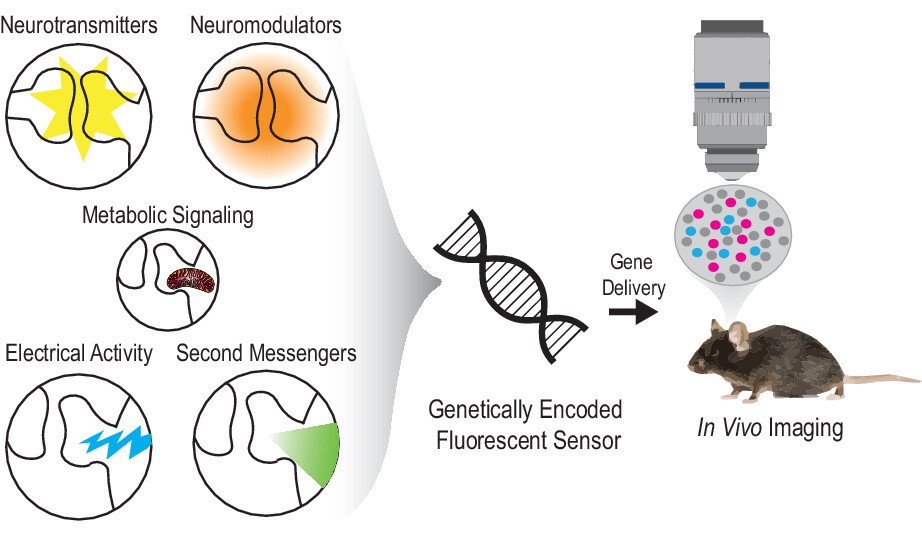
Check out our review of fluorescent sensors in the Journal of Neurochemistry
Genetically encoded fluorescent sensors for imaging neuronal dynamics in vivo.
↗ 10.1111/jnc.15608

Get proficient with CRISPIE following our new Bio-Protocol
Labeling Endogenous Proteins Using CRISPR-mediated Insertion of Exon (CRISPIE).
↗ 10.21769/BioProtoc.4343
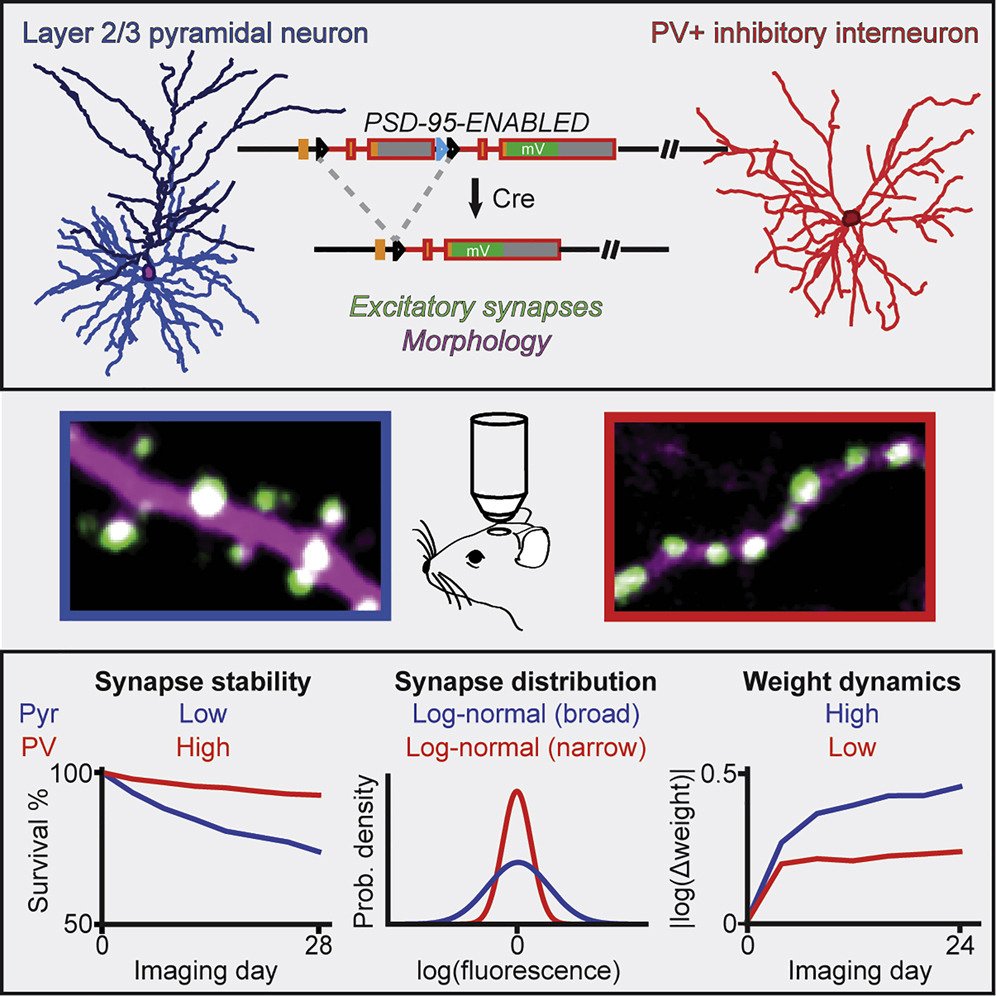
Read our Cell Reports paper comparing synaptic properties of PV-interneurons and pyramidal neurons using endogenous labeling of PSD-95
Distinct in vivo dynamics of excitatory synapses onto cortical pyramidal neurons and parvalbumin-positive interneurons.
↗ 10.1016/j.celrep.2021.109972

Read our new review of genetically encoded in vivo cAMP and PKA sensors in Journal of Neuroscience Methods
Genetically encoded sensors towards imaging cAMP and PKA activity in vivo.
↗ 10.1016/j.jneumeth.2021.109298
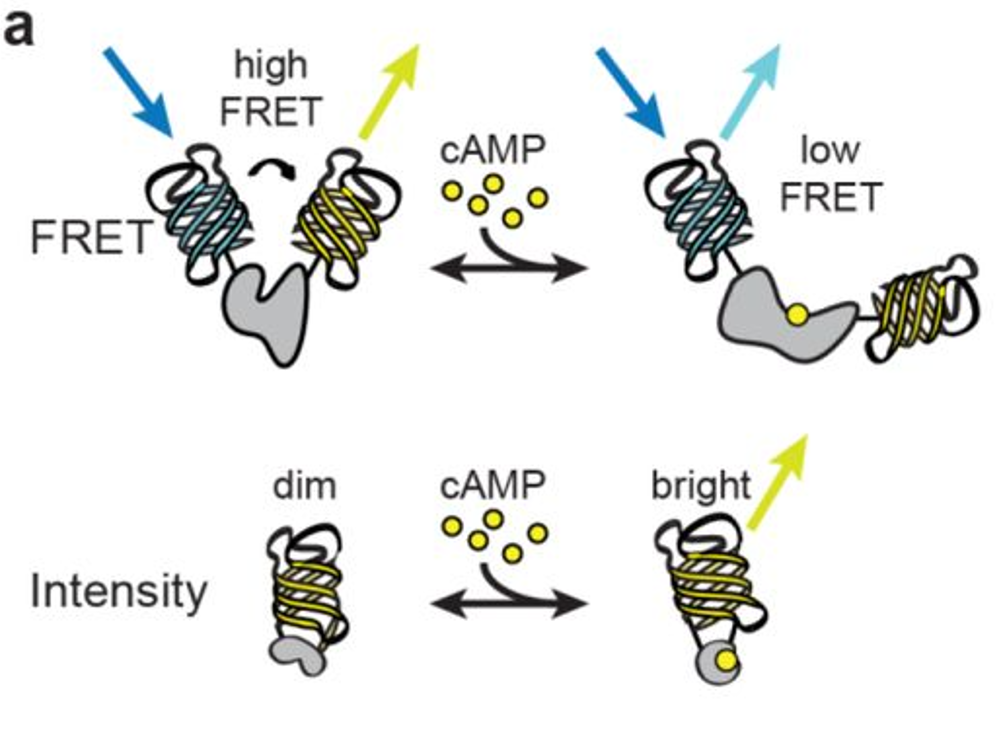
Our cAMP sensor, cAMPFIRE, is out in BioRxiv!
Highly sensitive genetically-encoded sensors for population and subcellular imaging of cAMP in vivo.
↗ 10.7554/eLife.64911

Read our paper in eLife introducing a new method for designer exon insertion: CRISPIE
High-fidelity, efficient, and reversible labeling of endogenous proteins using CRISPR-based designer exon insertion.
↗ 10.7554/eLife.64911




